100 өөрчлөгдсөн 55 нэмэгдсэн , 35 устгасан
+ 2
- 1
.gitignore
|
||
|
||
|
||
|
||
|
||
|
||
Файлын зөрүү хэтэрхий том тул дарагдсан байна
+ 9
- 7
approach1a_basic_frame_differencing.ipynb
Файлын зөрүү хэтэрхий том тул дарагдсан байна
+ 12
- 9
approach2_background_estimation.ipynb
BIN
approach3_dsift.png

BIN
approach3_keypoints.pdf
BIN
approach3_keypoints_lapse.pdf
Файлын зөрүү хэтэрхий том тул дарагдсан байна
+ 0
- 0
approach3_local_features.ipynb
Файлын зөрүү хэтэрхий том тул дарагдсан байна
+ 6
- 6
approach4_autoencoder.ipynb
BIN
approach4_difficult_anomalous_beaver_01.png

BIN
approach4_difficult_anomalous_marten_01.png

BIN
approach4_difficult_normal_beaver_01.png

BIN
approach4_difficult_normal_marten_01.png

BIN
approach4_easy_anomalous_beaver_01.png

BIN
approach4_easy_anomalous_marten_01.png

BIN
approach4_easy_normal_beaver_01.png

BIN
approach4_easy_normal_marten_01.png

+ 10
- 5
eval_autoencoder.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 16
- 7
eval_bow.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
imgs/approach1a_lapse.pdf
BIN
imgs/approach1a_motion.pdf
BIN
imgs/approach1a_sigma4_lapse.pdf
BIN
imgs/approach1a_sigma4_motion.pdf
BIN
imgs/approach1a_sigma4_sqdiff.pdf
BIN
imgs/approach1a_sqdiff.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma6.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma6.png

BIN
plots/approach1a/roc_curves/Beaver_01_absvar.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma4.png
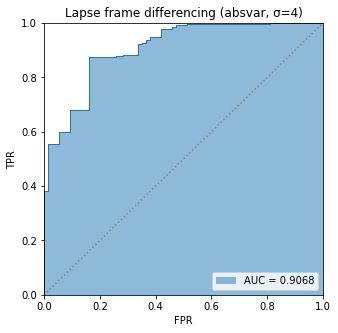
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma6.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma6.png
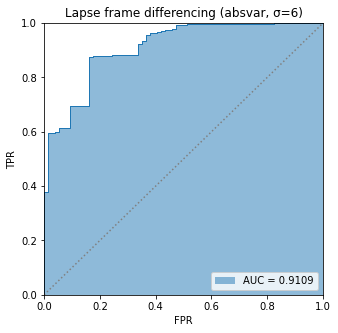
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma6.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma6.png

BIN
plots/approach1a/roc_curves/Beaver_01_sqvar.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma6.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma6.png

BIN
plots/approach2/roc_curves/Beaver_01_sqmean.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean.png
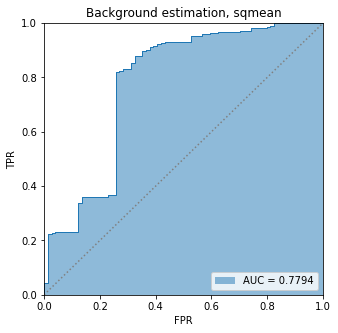
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma2.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma2.png
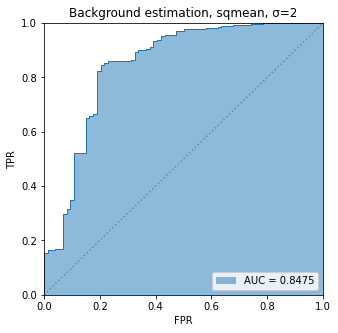
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma4.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma4.png

BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma6.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma6.png

BIN
plots/approach2/roc_curves/Beaver_01_sqvar.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar.png

BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma2.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma2.png

BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma4.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma4.png
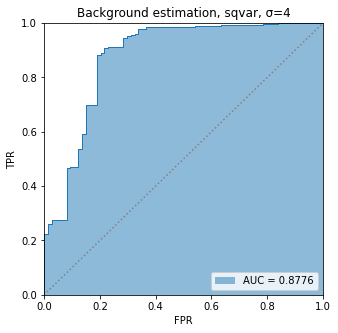
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma6.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma6.png

BIN
plots/approach3/roc_curves/Beaver_01_30_30_1024.pdf
BIN
plots/approach3/roc_curves/Beaver_01_30_30_1024.png

BIN
plots/approach4/roc_curves/Beaver_01_kde.pdf
BIN
plots/approach4/roc_curves/Beaver_01_kde.png

BIN
plots/approach4/roc_curves/Beaver_01_loss.pdf
BIN
plots/approach4/roc_curves/Beaver_01_loss.png
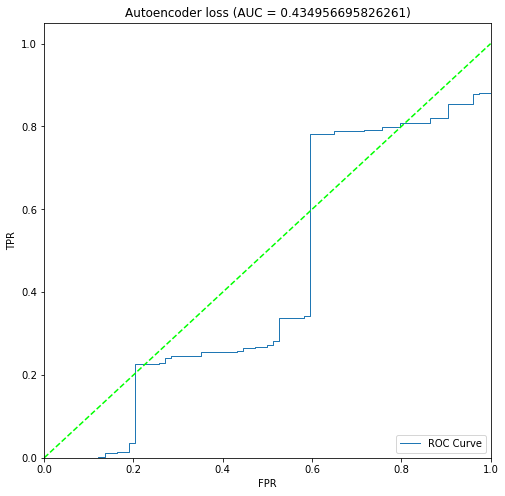
BIN
plots/approach4/roc_curves/GFox_03_kde,loss.pdf
BIN
plots/approach4/roc_curves/GFox_03_kde,loss.png

BIN
plots/approach4/roc_curves/GFox_03_kde.pdf
BIN
plots/approach4/roc_curves/GFox_03_kde.png

BIN
plots/approach4/roc_curves/GFox_03_loss.pdf
BIN
plots/approach4/roc_curves/GFox_03_loss.png

BIN
plots/approach4/roc_curves/Marten_01_kde,loss.pdf
BIN
plots/approach4/roc_curves/Marten_01_kde,loss.png

BIN
plots/approach4/roc_curves/Marten_01_kde.pdf
BIN
plots/approach4/roc_curves/Marten_01_loss.pdf
BIN
plots/approach4/roc_curves/Marten_01_loss.png

BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_Beaver_01_kde.pdf
BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_Beaver_01_kde.png

BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_Beaver_01_loss.pdf
BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_Beaver_01_loss.png

BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_exp_Beaver_01_kde.pdf
BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_exp_Beaver_01_kde.png

BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_nodo_Beaver_01_kde.pdf
BIN
plots/approach4/roc_curves/ae2_deep_beaver_01_nodo_Beaver_01_kde.png

BIN
plots/approach4/roc_curves/ae2_deep_gfox_03_GFox_03_kde.pdf
BIN
plots/approach4/roc_curves/ae2_deep_gfox_03_GFox_03_kde.png

BIN
plots/approach4/roc_curves/ae2_deep_gfox_03_GFox_03_loss.pdf
BIN
plots/approach4/roc_curves/ae2_deep_gfox_03_GFox_03_loss.png

BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_kde,loss.pdf
BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_kde,loss.png

BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_kde.pdf
BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_kde.png

BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_loss.pdf
BIN
plots/approach4/roc_curves/ae2_deep_marten_01_Marten_01_loss.png
