80 changed files with 194 additions and 78 deletions
File diff suppressed because it is too large
+ 34
- 12
approach1a_basic_frame_differencing.ipynb
File diff suppressed because it is too large
+ 8
- 11
approach1b_histograms.ipynb
File diff suppressed because it is too large
+ 10
- 9
approach2_background_estimation.ipynb
File diff suppressed because it is too large
+ 5
- 13
approach3_local_features.ipynb
+ 77
- 0
day_vs_night.ipynb
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 10
- 4
eval_bow.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
plots/approach1a/roc_curves/Beaver_01.png
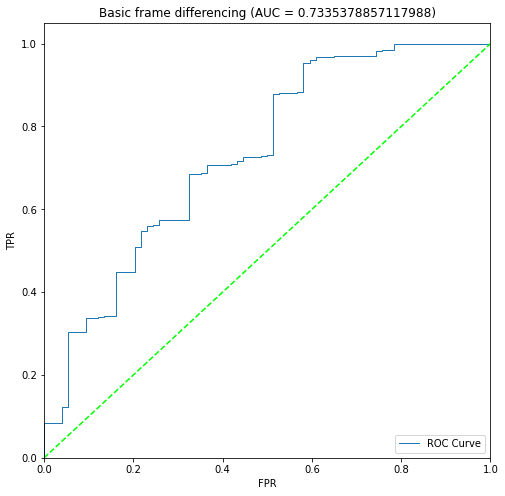
BIN
plots/approach1a/roc_curves/Beaver_01_absmean.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean.png

BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma2.png

BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absmean_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_absstd.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absstd.png
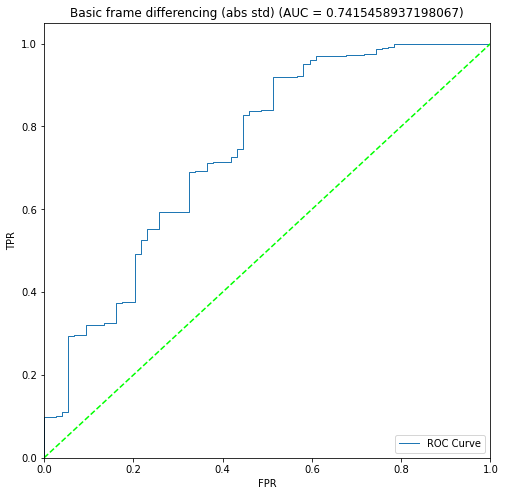
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma2.png
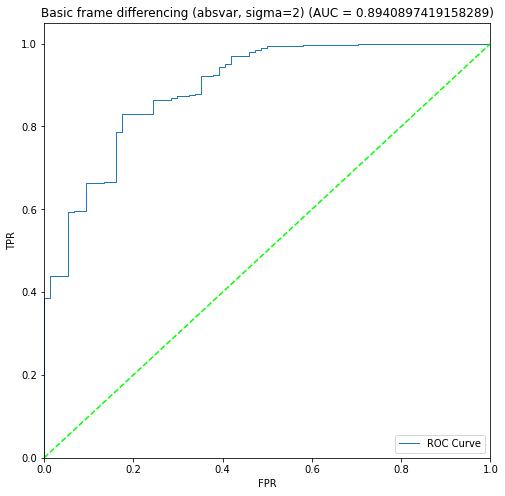
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_absvar_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sigma2.png
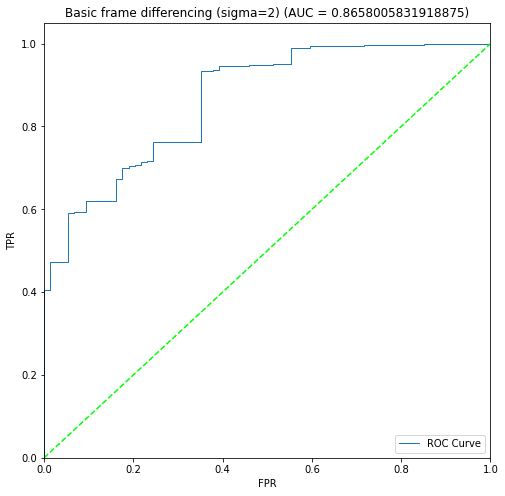
BIN
plots/approach1a/roc_curves/Beaver_01_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sigma4.pdf → plots/approach1a/roc_curves/Beaver_01_sqmean.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean.png
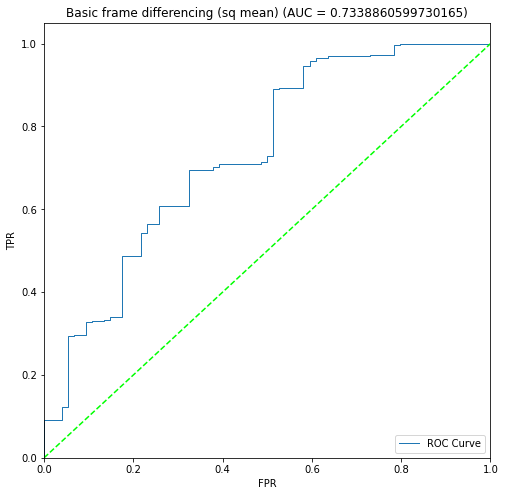
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma2.png

BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqmean_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sqvar.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar.png
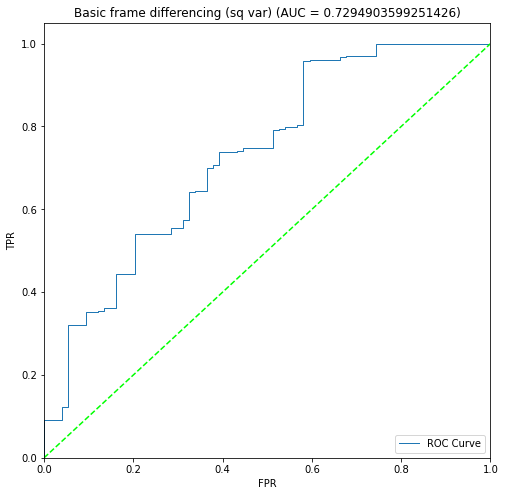
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma2.png
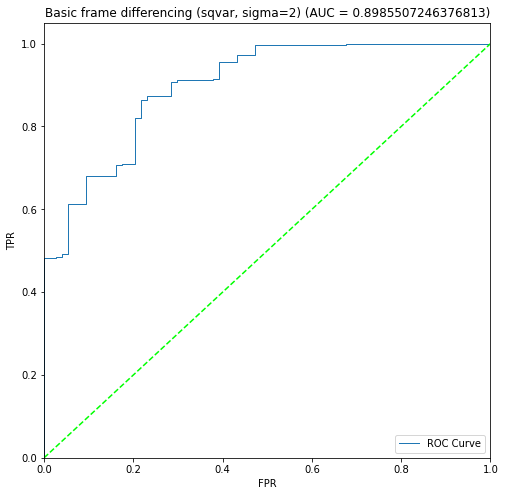
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Beaver_01_sqvar_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01.pdf → plots/approach1a/roc_curves/Marten_01_absmean.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absmean.png

BIN
plots/approach1a/roc_curves/Marten_01_absmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absmean_sigma2.png

BIN
plots/approach1a/roc_curves/Marten_01_absmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absmean_sigma4.png
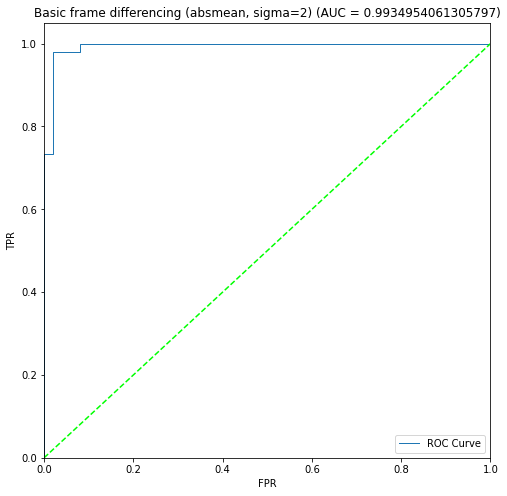
BIN
plots/approach1a/roc_curves/Marten_01_absvar.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absvar.png

BIN
plots/approach1a/roc_curves/Marten_01_absvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absvar_sigma2.png

BIN
plots/approach1a/roc_curves/Marten_01_absvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Marten_01_absvar_sigma4.png

BIN
plots/approach1a/roc_curves/Beaver_01_sigma2.pdf → plots/approach1a/roc_curves/Marten_01_sqmean.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqmean.png

BIN
plots/approach1a/roc_curves/Marten_01_sqmean_sigma2.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqmean_sigma2.png
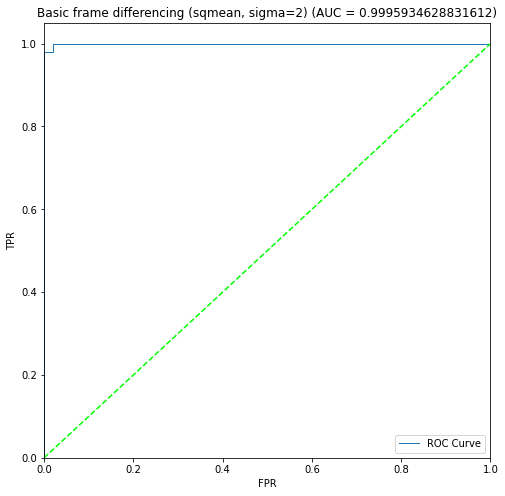
BIN
plots/approach1a/roc_curves/Marten_01_sqmean_sigma4.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqmean_sigma4.png
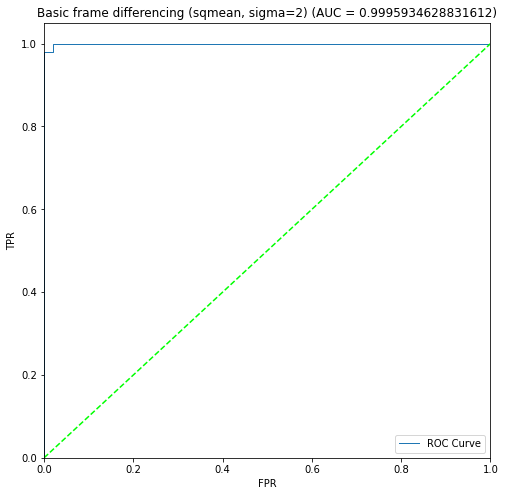
BIN
plots/approach1a/roc_curves/Marten_01_sqvar.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqvar.png

BIN
plots/approach1a/roc_curves/Marten_01_sqvar_sigma2.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqvar_sigma2.png

BIN
plots/approach1a/roc_curves/Marten_01_sqvar_sigma4.pdf
BIN
plots/approach1a/roc_curves/Marten_01_sqvar_sigma4.png

BIN
plots/approach1b/roc_curves/Beaver_01_pmean.pdf
BIN
plots/approach1b/roc_curves/Beaver_01_pmean.png
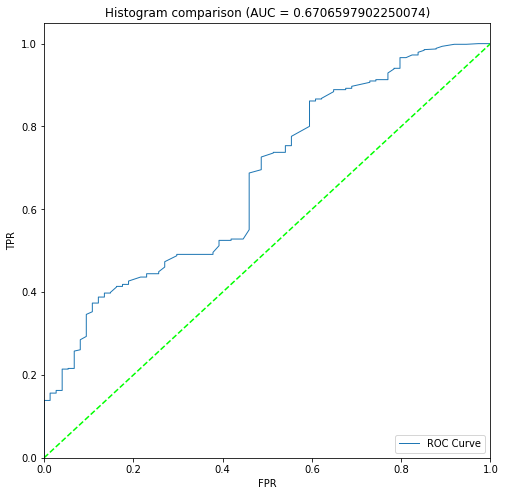
BIN
plots/approach2/roc_curves/Beaver_01_sqmean.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean.png
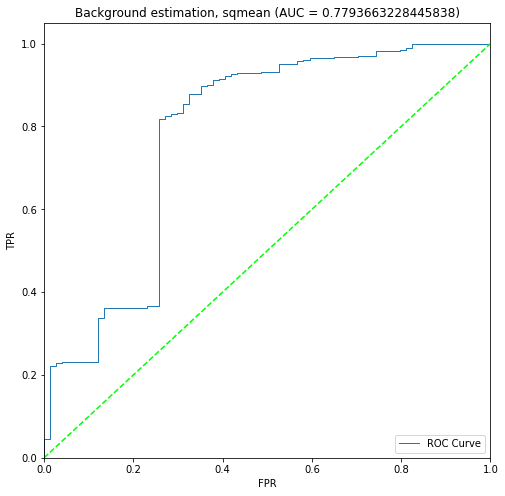
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma2.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma2.png

BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma4.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma4.png

BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma6.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqmean_sigma6.png

BIN
plots/approach2/roc_curves/Beaver_01_sqvar.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar.png

BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma2.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma2.png
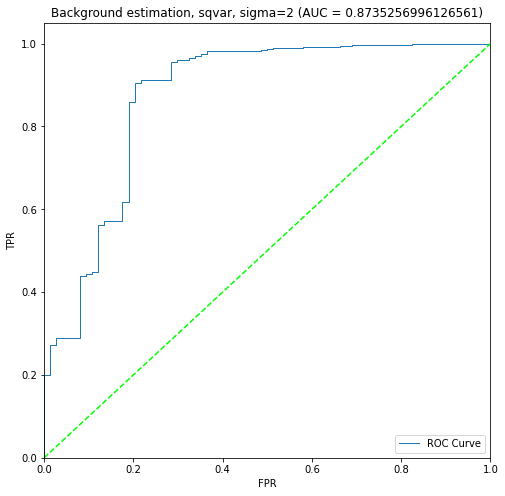
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma4.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma4.png
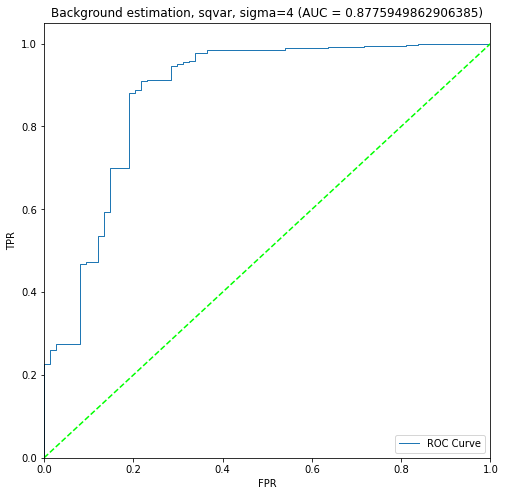
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma6.pdf
BIN
plots/approach2/roc_curves/Beaver_01_sqvar_sigma6.png
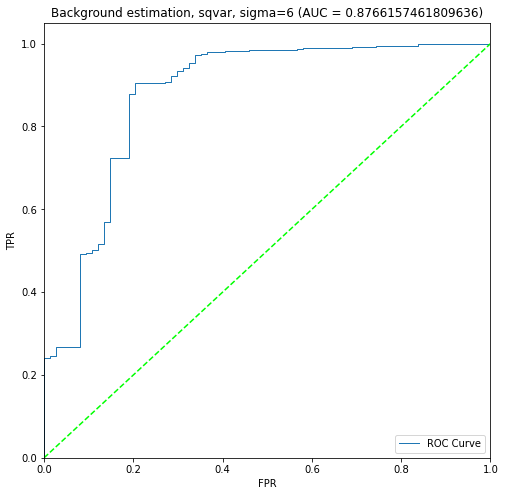
+ 3
- 1
py/ImageUtils.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 10
- 7
py/LocalFeatures.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 11
- 3
py/PlotUtils.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 20
- 13
results.ipynb
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 6
- 5
train_bow.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||