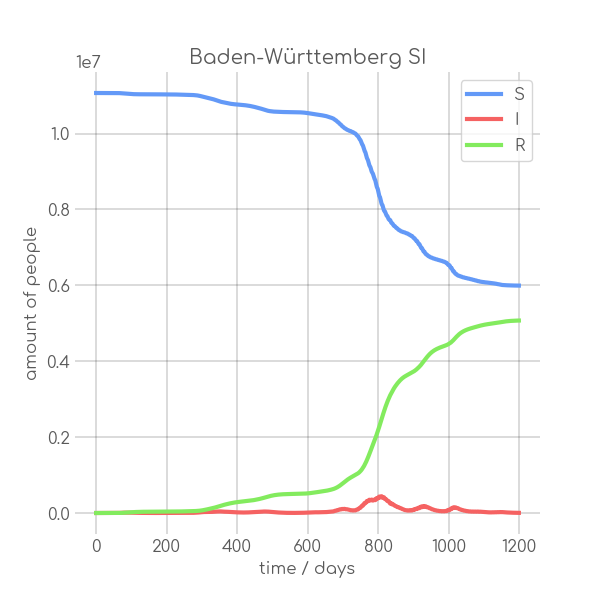
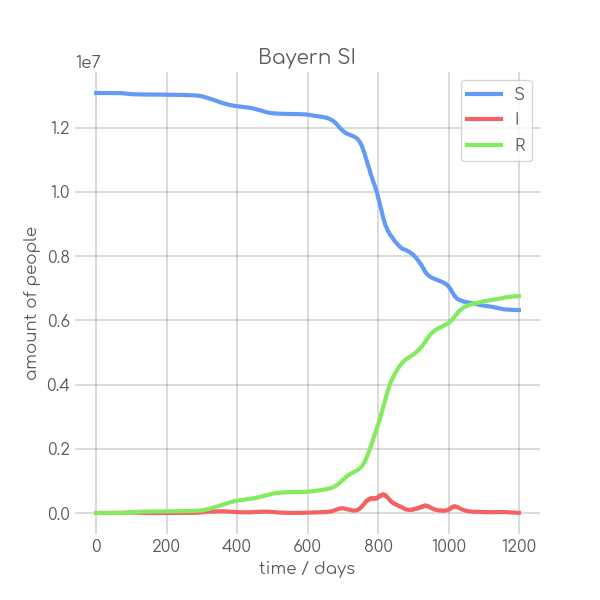
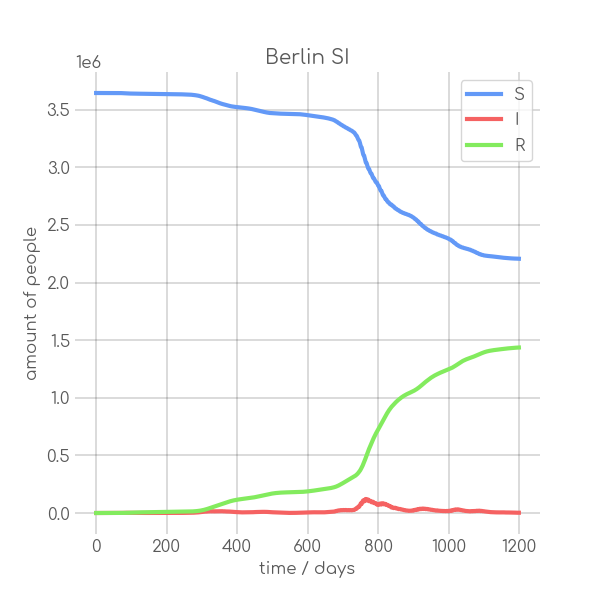
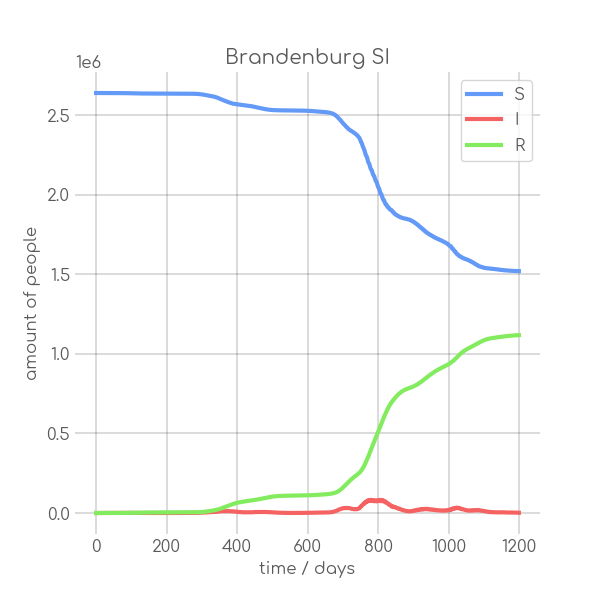
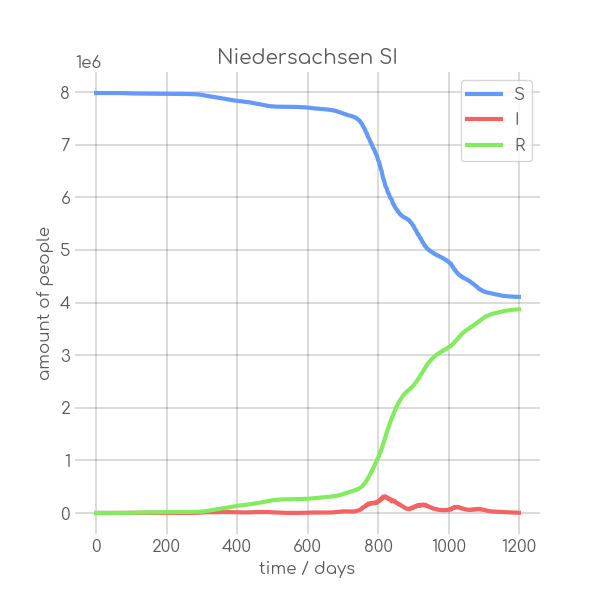
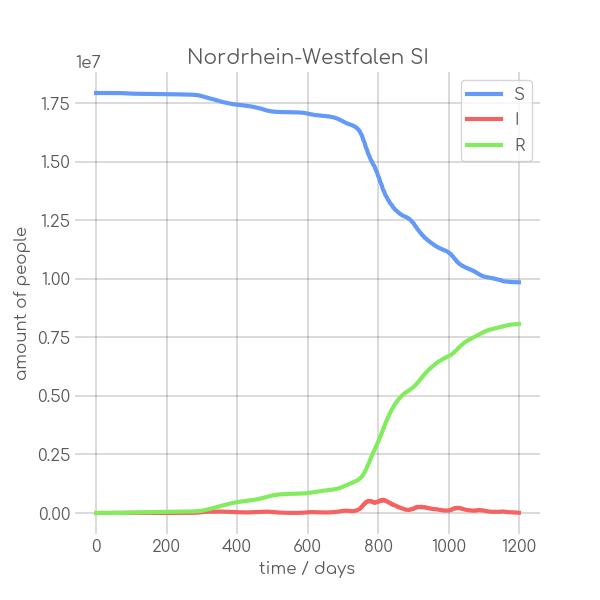
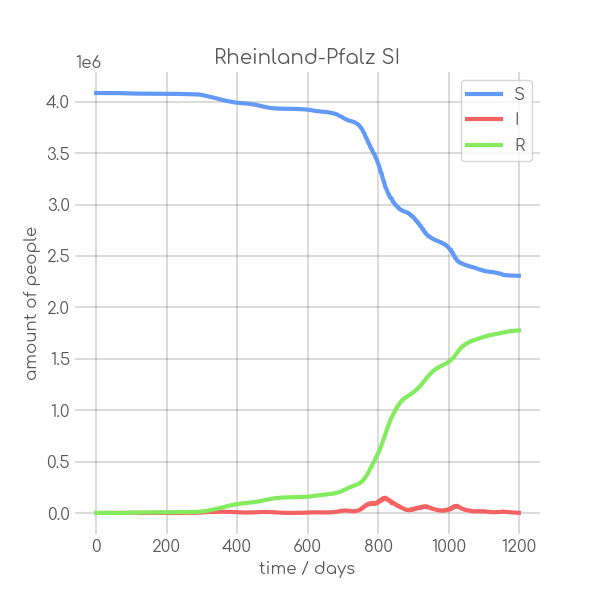
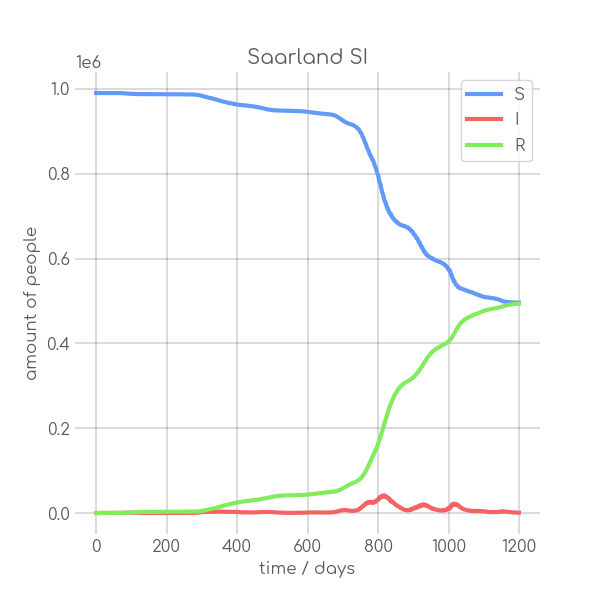
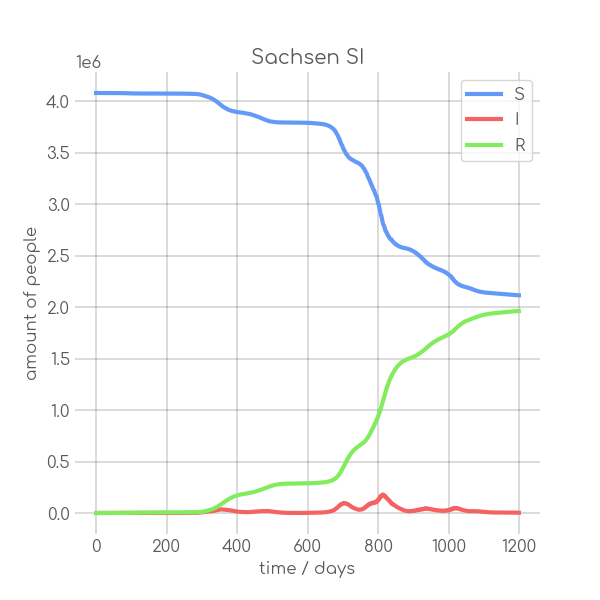
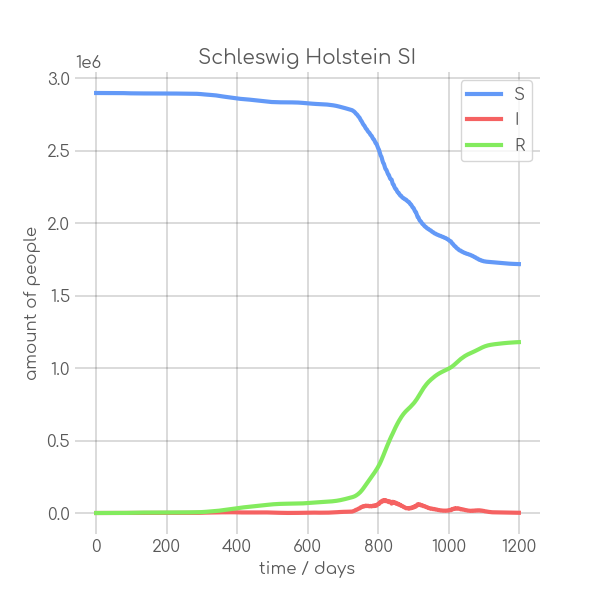
|
|
@@ -86,7 +86,7 @@ def get_state_cases(county_id, state_id):
|
|
|
id = county_id // 1000
|
|
|
return id == state_id
|
|
|
|
|
|
-def state_based_data(plotter:Plotter, state_name:str, time_range=1200, sample_rate=1, dataset_path='datasets/state_data/Aktuell_Deutschland_SarsCov2_Infektionen.csv'):
|
|
|
+def state_based_data(plotter:Plotter, state_name:str, model='SIR', alpha=1/14, time_range=1200, sample_rate=1, dataset_path='datasets/state_data/Aktuell_Deutschland_SarsCov2_Infektionen.csv'):
|
|
|
"""Transforms the RKI infection cases dataset to a SIR dataset.
|
|
|
|
|
|
Args:
|
|
|
@@ -147,23 +147,40 @@ def state_based_data(plotter:Plotter, state_name:str, time_range=1200, sample_ra
|
|
|
|
|
|
S = np.zeros(time_range)
|
|
|
I = np.zeros(time_range)
|
|
|
+ R = np.zeros(time_range)
|
|
|
+
|
|
|
+ # generate groups
|
|
|
S[0] = N - infections[0]
|
|
|
I[0] = infections[0]
|
|
|
+ R[0] = 0
|
|
|
|
|
|
for day in range(1, time_range):
|
|
|
S[day] = S[day-1] - infections[day]
|
|
|
- I[day] = I[day-1] + infections[day] - I[day-1]/14
|
|
|
+ I[day] = I[day-1] + infections[day] - I[day-1] * alpha
|
|
|
+ R[day] = R[day-1] + I[day-1] * alpha
|
|
|
|
|
|
t = np.arange(0, time_range, 1)
|
|
|
|
|
|
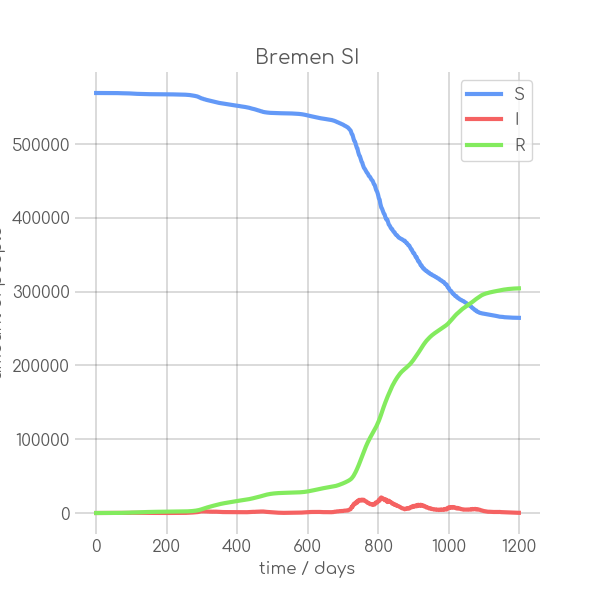
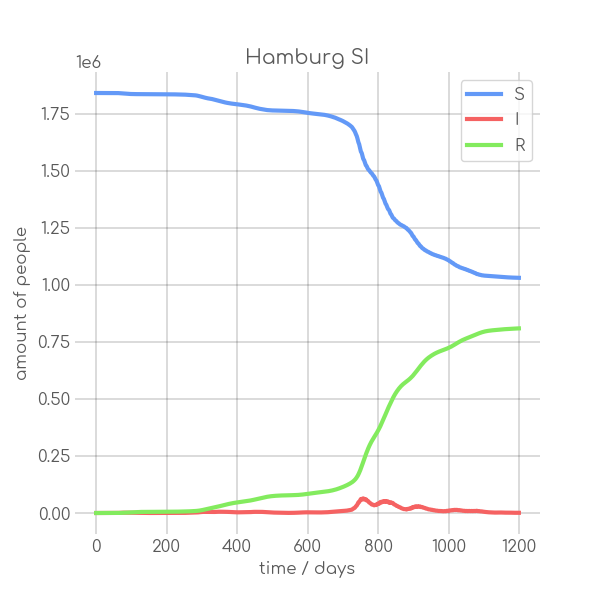
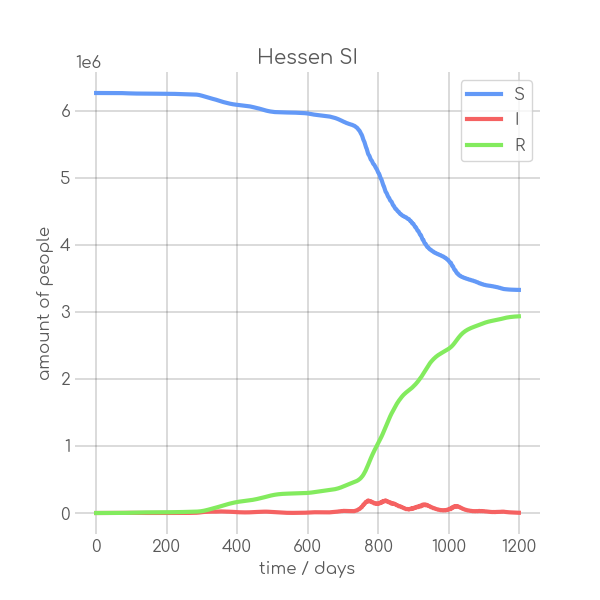
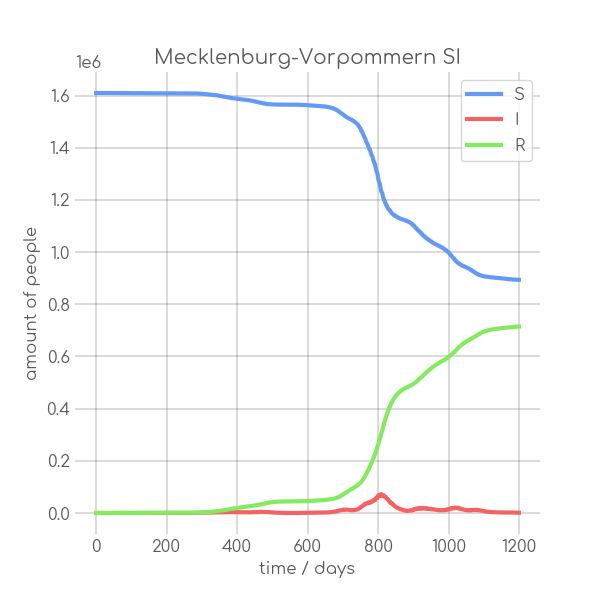
- plotter.plot(t, [S, I], ['S', 'I'], state_name.replace(' ', '_').replace('-', '_').replace('ü','ue'), state_name+' SI', (6,6), xlabel='time / days', ylabel='amount of people')
|
|
|
-
|
|
|
- COVID_Data = np.asarray([t[0::sample_rate],
|
|
|
- S[0::sample_rate],
|
|
|
- I[0::sample_rate]])
|
|
|
+ # select, which group is to be outputted
|
|
|
+ groups = []
|
|
|
+ if 'S' in model:
|
|
|
+ groups.append(S)
|
|
|
+
|
|
|
+ if 'I' in model:
|
|
|
+ groups.append(I)
|
|
|
|
|
|
- np.savetxt(f"datasets/SIR_RKI_{state_name.replace(' ', '_').replace('-', '_').replace('ü','ue')}_{sample_rate}.csv", COVID_Data, delimiter=",")
|
|
|
+ if 'R' in model:
|
|
|
+ groups.append(R)
|
|
|
|
|
|
+ plotter.plot(t,
|
|
|
+ groups,
|
|
|
+ [*model],
|
|
|
+ state_name.replace(' ', '_').replace('-', '_').replace('ü','ue'),
|
|
|
+ state_name +' SI',
|
|
|
+ (6,6),
|
|
|
+ xlabel='time / days',
|
|
|
+ ylabel='amount of people')
|
|
|
|
|
|
+ COVID_Data = np.asarray([t[0::sample_rate]] + [group[0::sample_rate] for group in groups])
|
|
|
|
|
|
-
|
|
|
+ np.savetxt(f"datasets/{model}_RKI_{state_name.replace(' ', '_').replace('-', '_').replace('ü','ue')}_{sample_rate}.csv", COVID_Data, delimiter=",")
|