|
|
@@ -40,7 +40,7 @@ class SyntheticDeseaseData:
|
|
|
if self.generated:
|
|
|
self.plotter.plot(self.t, self.data, labels, title, title, (6, 6), xlabel='time / days', ylabel='amount of people')
|
|
|
else:
|
|
|
- print('Data has to be generated before plotting!') # Fabienne war hier
|
|
|
+ print('Data has to be generated before plotting!')
|
|
|
|
|
|
class SI(SyntheticDeseaseData):
|
|
|
def __init__(self, plotter:Plotter, N=59e6, I_0=1, simulation_time=500, time_points=100, alpha=0.191, beta=0.05) -> None:
|
|
|
@@ -102,6 +102,45 @@ class SI(SyntheticDeseaseData):
|
|
|
else:
|
|
|
print('Data has to be generated before plotting!')
|
|
|
|
|
|
+class I(SI):
|
|
|
+ def __init__(self, plotter:Plotter, N=59e6, I_0=1, simulation_time=500, time_points=100, alpha=0.191, beta=0.05) -> None:
|
|
|
+ """This class is able to generate synthetic data of the SI groups for the reduced SIR model. This is done by utiling the SIR model.
|
|
|
+
|
|
|
+ Args:
|
|
|
+ plotter (Plotter): Plotter object to plot dataset curves.
|
|
|
+ N (int, optional): Size of the population. Defaults to 59e6.
|
|
|
+ I_0 (int, optional): Initial size of the infectious group. Defaults to 1.
|
|
|
+ simulation_time (int, optional): Real time for that the synthetic data is supposed to be generated in days. Defaults to 500.
|
|
|
+ time_points (int, optional): Number of time sample points. Defaults to 100.
|
|
|
+ alpha (float, optional): Factor dictating how many people per timestep go from 'Infectious' to 'Removed'. Defaults to 0.191.
|
|
|
+ beta (float, optional): Factor dictating how many people per timestep go from 'Susceptible' to 'Infectious'. Defaults to 0.05.
|
|
|
+ """
|
|
|
+
|
|
|
+ super().__init__(plotter, N=N, I_0=I_0, simulation_time=simulation_time, time_points=time_points, alpha=alpha, beta=beta)
|
|
|
+
|
|
|
+ def generate(self):
|
|
|
+ """This funtion generates the data for this configuration of the SIR model.
|
|
|
+ """
|
|
|
+ super().generate()
|
|
|
+ self.data = self.data[1]
|
|
|
+ print(self.data.shape)
|
|
|
+
|
|
|
+ def plot(self, title=''):
|
|
|
+ """Plot the data which was generated.
|
|
|
+ """
|
|
|
+ if self.generated:
|
|
|
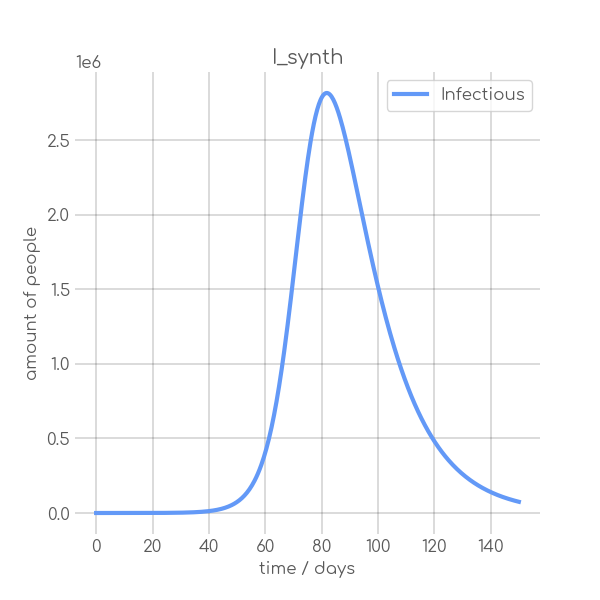
+ self.plotter.plot(self.t, [self.data], ['Infectious'], title, title, (6, 6), xlabel='time / days', ylabel='amount of people')
|
|
|
+ else:
|
|
|
+ print('Data has to be generated before plotting!')
|
|
|
+
|
|
|
+ def save(self, name=''):
|
|
|
+ if self.generated:
|
|
|
+ COVID_Data = np.asarray([self.t, self.data])
|
|
|
+
|
|
|
+ np.savetxt('datasets/I_data.csv', COVID_Data, delimiter=",")
|
|
|
+ else:
|
|
|
+ print('Data has to be generated before plotting!')
|
|
|
+
|
|
|
|
|
|
class SIR(SyntheticDeseaseData):
|
|
|
def __init__(self, plotter:Plotter, N=59e6, I_0=1, R_0=0, simulation_time=500, time_points=100, alpha=0.191, beta=0.05) -> None:
|